
The FPsc/R500 (molecular) genetic map
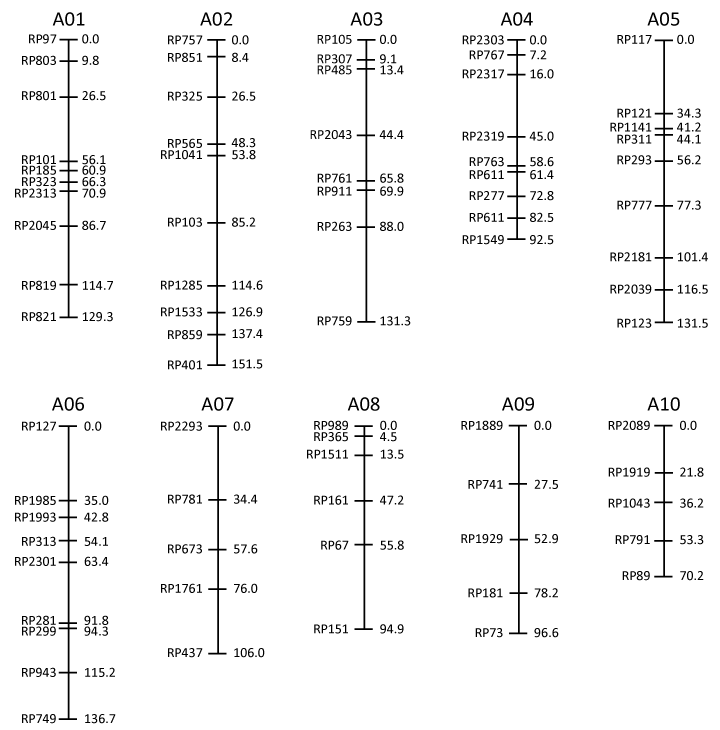
Students in our lab group at UW-Madison have worked hard over the past several years to generate a collection of polymorphic molecular markers useful for mapping FPsc mutant loci. The current map, shown below, provides marker names, chromosome assignments, and linkage relationships among marker loci as determined analysis of ~145 F2 segregants derived from an initial cross between the wild-type FPsc variety and an alternative B. rapa variety known as R500.
Primer sequences tested in the course of the FPsc marker development program were obtained from the published literature as cited in footnote 2 of the excel spreadsheet or created de novo by using BAC sequences and, later, the whole genome sequence of B. rapa variety Chiifu to identify hypermutable simple sequence repeats. Throughout, the primary criteria used to evaluate marker quality were (1) that the primer sequences amplified robustly from FPsc, R500, and FPsc/R500 heterozygote gDNA templates using a single and rapid thermocycling profile, and (2) that the size difference of PCR products produced from alternative alleles were sufficient to permit unambiguous genotypic assignments.
Marker assays were conducted through PCRs that use a common thermocycling profile (35 cycles at 54 degr. C annealing temperature) and reaction products were visualized by electrophoresis through 2% agarose gels. Experimental protocols for genomic DNA extractions, PCRs and gel electrophoresis are available here. Linkage relationships among marker loci were determined by using the R/QTL software package, ably assisted by Karl Broman, UW-Madison. Oligonucleotide primer sequences and banding patterns observed for FPsc, FPsc/R500, and R500 genotypes are available in Adobe PDF and Microsoft Excel spreadsheet formats by clicking on the highlighted links.
Disclaimer: The FPsc/R500 genetic map shown here, like all scientific results, is presented as a tentative conclusion that is subject to change as additional data may warrant.
